Research
Our lab broadly investigates how biochemical and mechanical signals at cell-cell and cell-extracellular matrix adhesions are coordinated across biological scales (molecules to cells to tissues) to maintain normal tissue structure or drive pathology. Our efforts are primarily focused in the experimental arenas of cardiovascular disease and epithelial growth and development.
NEW INSIGHTS INTO Notch Receptor MEchanobiology and Function
|
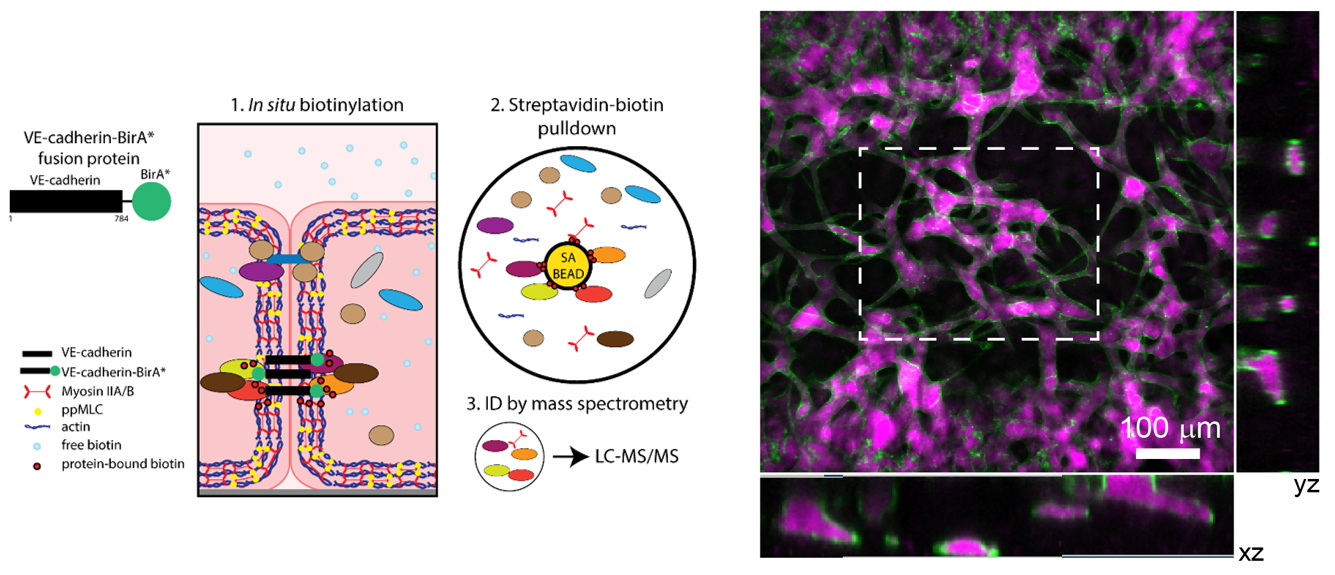
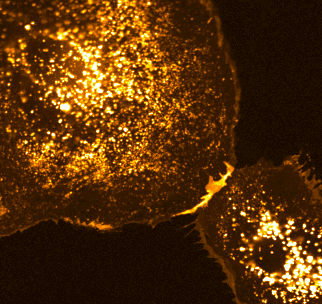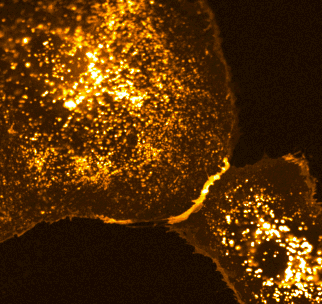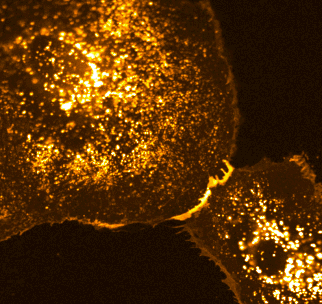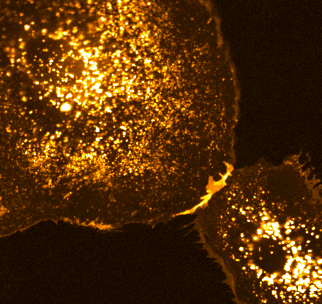
Two families of proteins that operate at the interface of cell-cell contact, Notch and cadherin receptors, are essential for metazoan multicellularity. Notch receptors are ubiquitously important for cellular fate decisions and function as transcriptional activators in many cells and tissues throughout development, homeostasis and disease, while cadherin receptors mechanically stabilize cell-cell interactions and establish organized nexuses of signaling activity within the plasma membrane. Intriguingly, we have identified a previously undescribed cortical signaling pathway by which Notch receptor activation results in both canonical transcriptional activity and the stability of cadherin receptors and the cortical actin cytoskeleton, providing a new mechanism by which a single receptor might link transcriptional programs with adhesive and cytoskeletal remodeling.
|
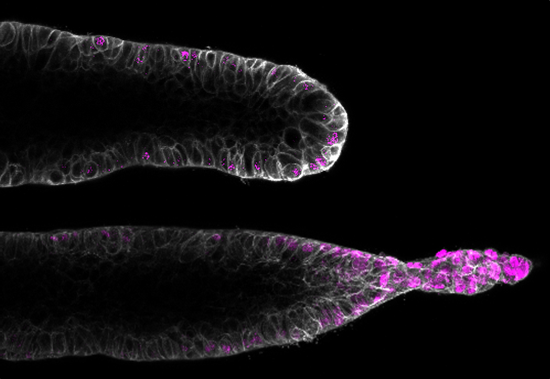
Orchestration of vascular morphogenesis by cell-cell adhesions
|
Current project focuses:
Mechanisms of endothelial shear stress mechanotransduction and vascular barrier function. Integration of inflammatory and mitogenic signaling at endothelial cell-cell adhesions. Understanding angiogenesis and mechanisms of endothelial symmetry breaking. Modeling and dissecting vascular malformations. |
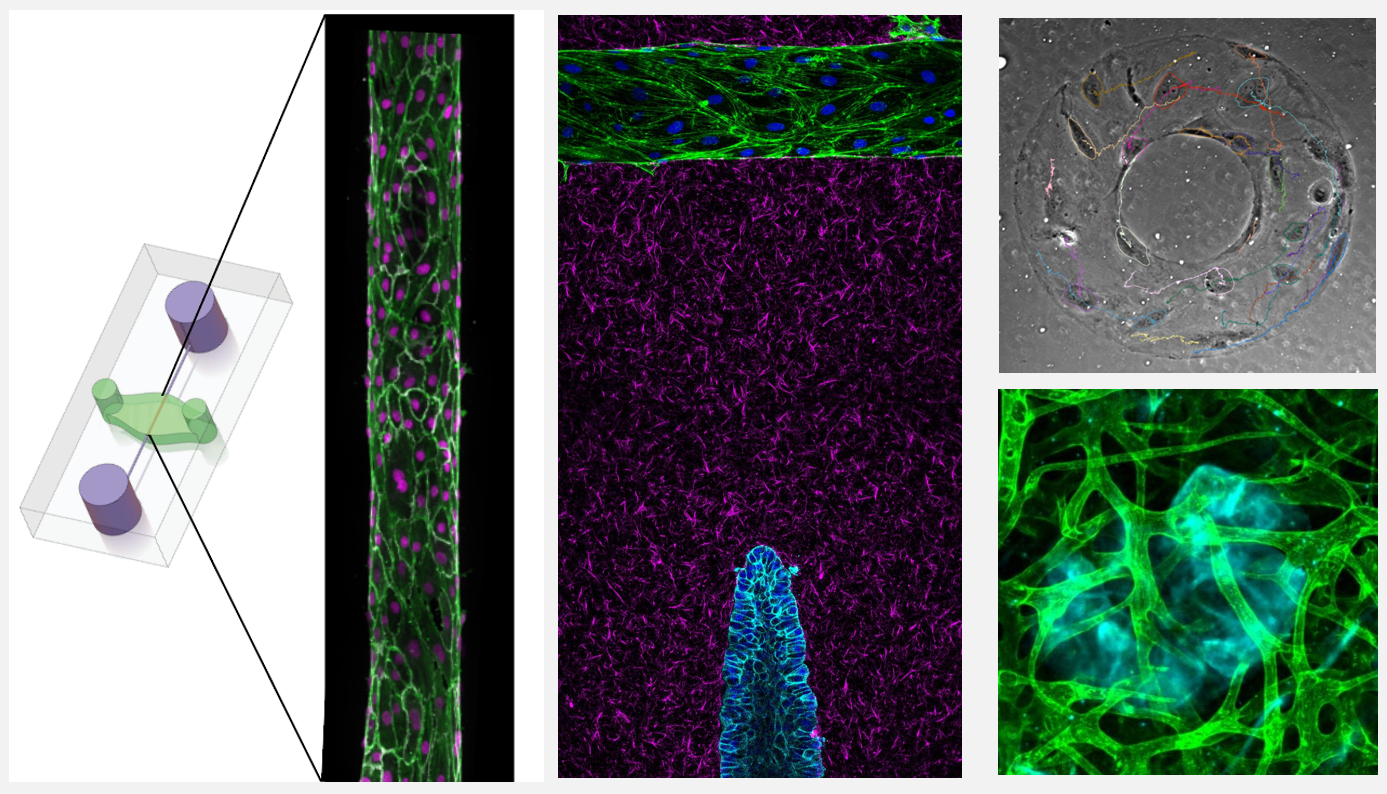
Human biomimetic systems: discovery platforms for tissue morphogenesis and mechanobiology
|
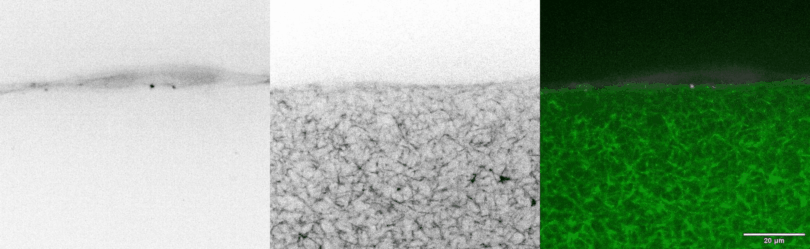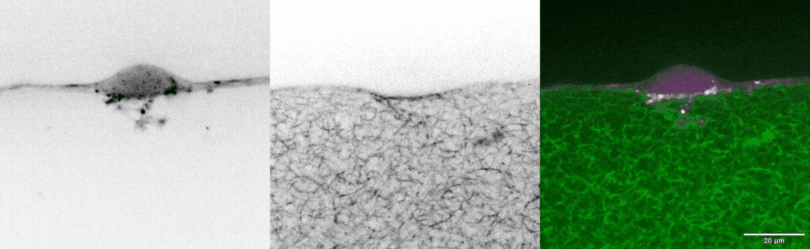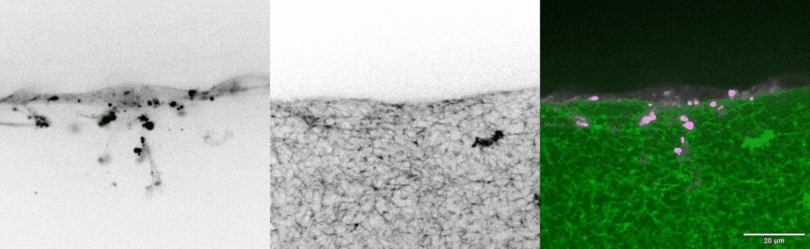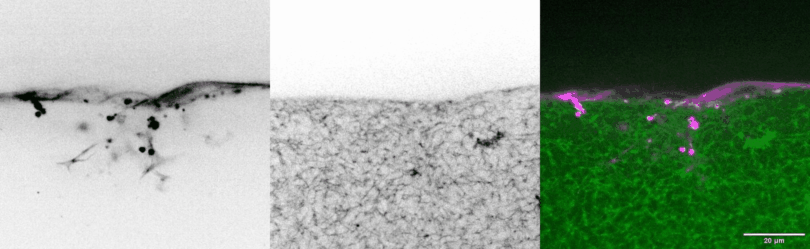
In vivo models are the standard for studying tissue morphogenesis and its regulatory signaling, largely due to a lack of human in vitro systems that recapitulate key structural features, microenvironmental cues, and morphogenic behaviors of in vivo normal and transformed tissues. To address this experimental need, we have engineered biomimetic systems that model 3D human tissues of organotypic architectures in vitro, and permit the simulation, molecular interrogation, and quantitative analysis of in vivo-like morphogenic processes. Integrating these platforms with classic cell biological approaches and cellular and molecular technologies has enabled multiple discoveries of previously undescribed morphogenic and mechanobiological signaling pathways.
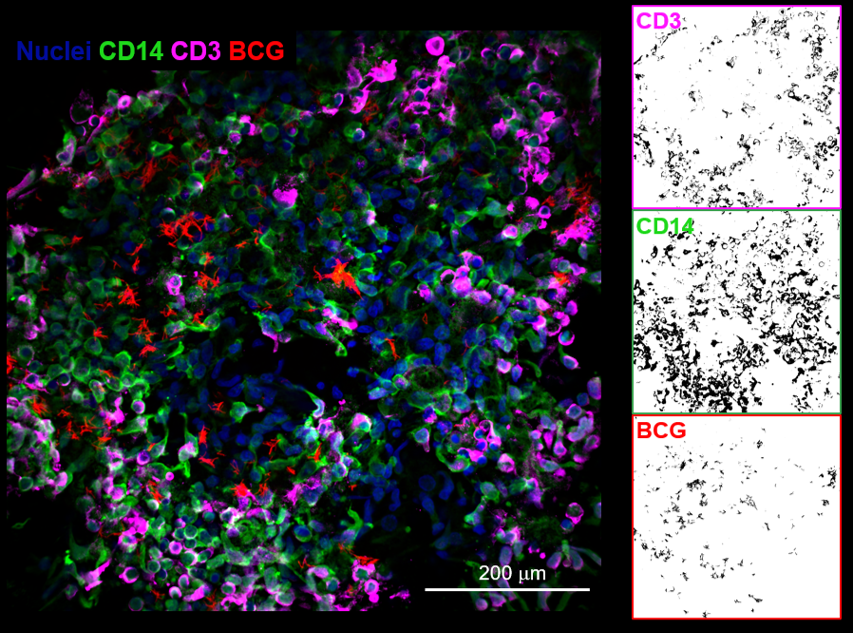
In an exciting application of this technology, we have developed biomimetic human tuberculosis granulomas in collaboration with Dr. Joel Ernst. We are employing this platform to uncover fundamental new mechanisms relating stromal signaling to host immune responses in disease pathogenesis. |